The fourth iteration of our
fragment-finding methods poll has just closed. If you want to jump right to the
results feel free to skip the next paragraph, which focuses on methods.
The poll was run using Crowdsignal,
the successor to Polldaddy, and ran from 20 October through 30 November. This
free polling software tabulates total number of votes for a question but not
the number of individual respondents. To determine individual respondents, we
included a question on “workplace and practice.” Of the 137 individual
respondents to this question, 116 identified themselves as practicing FBLD, and
we assumed they also answered the second question. The overall number of
responses is slightly higher than in 2013 but a bit lower than in 2016.
Readership demographics have shifted
from previous years, with about two thirds of respondents hailing from
industry, up from just over half historically. The fraction of respondents who
actively practice FBLD is also up modestly, to 85%.
But the question probably of most
interest is on screening methods, summarized here.
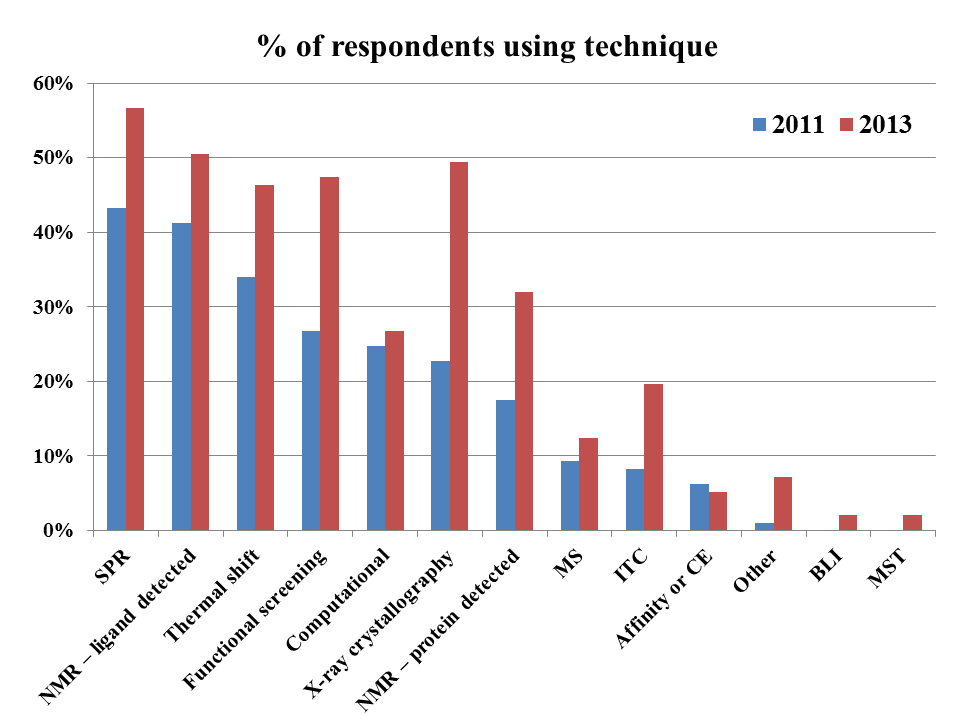
As we also saw in 2013 and 2016, nearly
all fragment-finding techniques are being used more, with the average
respondent employing 6 methods today compared with 4.1 in 2016, 3.6 in 2013,
and 2.4 in 2011.
X-ray crystallography has leapt
to first place, likely driven in part by increasing speed and automation as
well as by studies suggesting that crystallography can give impressively high hit rates.
As in 2016, ligand-detected NMR,
SPR, and thermal shift assays are all very popular. Use of computational
approaches has increased, though perhaps not as much as might be expected given
recent advances. Functional screening is the only technique for which use
has remained constant, or perhaps even declined very slightly from 2013.
For the first time we asked about
use of literature to identify fragments, and nearly a third of respondents said
they incorporate previously published fragments into their work. As the amount
of publicly available information continues to increase it will be interesting to
see whether this number grows.
More niche methods such as mass spectrometry, MST, affinity selection, and biolayer interferometry are gaining
adherents; 30 respondents reported using mass spectrometry, for example. While
fewer than 20% of respondents are using affinity chromatography (including WAC),
CE, or ultrafiltration, that proportion has nearly quadrupled from our previous
three polls, though we can’t say which of these related methods accounts for the
increase.
Finally, only four respondents reported
using “other” methods, such as SHG. Perhaps we’ll ask about this and other emerging methods explicitly next
time.
Do the results surprise you, or
are they consistent with what you are using at your organization?