Last week the CHI Drug Discovery
Chemistry (DDC)
meeting was held in San Diego. This was the largest ever, with more
than 900 participants, 95% of whom attended in person, up from 87% last year. I
won’t attempt to cover all fourteen tracks but will just touch on some of the
main themes.
Computational approaches
All four days of the conference
featured dedicated sessions on machine learning and artificial intelligence,
but since I was in other sessions I don’t know how relevant they were to FBLD.
If you attended an interesting talk please let me know so I can watch it
on-demand.
Among computational talks I
did
see, Antonina Nazarova (University of Southern California) provided an update
on V-SYNTHES, which we first wrote about
here. This synthon-based screening
approach now covers 36 billion molecules and has been tested against eight
different proteins, four of which yielded nanomolar hits when tested experimentally.
Computational methods have
historically treated proteins as rigid, though many targets are anything but.
Diane Joseph-McCarthy (Boston University) described an improvement to the pocket
finding approach
FTMap, called FTMove, to incorporate molecular dynamics by
starting with an ensemble of different crystal structures. A further advance is
E-FTMap, which expands the number of virtual probes from 16 to 119 to more
finely assess ligandable sites.
Benjamin Walters (Genentech) described
using protein dynamics to find cryptic pockets using ESP, or Experimental
Structure Prediction. In this approach, experimental data from hydrogen-deuterium
exchange (
HDX) or chemical shift perturbations (
CSPs) are used to constrain multiple
parallel computational simulations, leading to better models than flexible
docking, even for weak fragments.
Experimental approaches
Protein-detected NMR was the
first practical fragment-finding method, and Steve Fesik (Vanderbilt) described
using
SAR by NMR to find fragments binding to the papain-like protease of
SARS-CoV-2. These have been advanced to molecules with nanomolar affinity and
activity in cell-based assays.
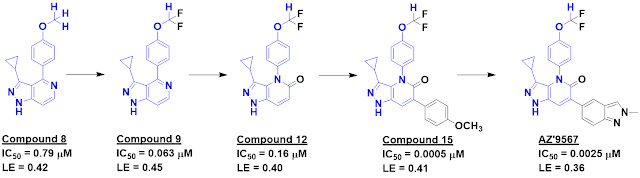
Andreas Lingel described the new fluorine-containing
fragment library built at Novartis and how
19F NMR was used to generate
inhibitors of IL-1β. We
wrote about that success last year, noting that the initial
fragment hit was “super-sized,” and Andreas confirmed that for trifluoromethyl-containing
fragments the upper molecular weight limit was relaxed to 350 Da.
Sriram Tyagarajan (Merck) presented
a crystallographic screen against the neurodegeneration target TTBK1 which yielded
hits at 15 sites. Several potential allosteric sites were identified, but
fragment growing and linking were not successful, leading them to a quick (3 month)
no-go decision on the protein.
Virgil Woods (City University of
New York) described using crystallographic screening to find hits against the challenging
phosphatase PTP1B both under conventional cryogenic temperatures as well as at
room temperature. As we
noted about related work, there was a surprisingly poor
overlap between the two sets of hits, and some fragments bound in a different
manner at different temperatures.
Integrating FBDD and DNA-encoded
libraries (DEL) for lead generation was the topic of Chaohong Sun’s talk. She
noted that of some two dozen targets at AbbVie screened by both methods, 60%
found hits from both, 10% found only fragment hits, and 5% found only DEL hits,
with a quarter of the targets producing no hits. Hits from both approaches can
be combined, as we noted
here. Chaohong also noted that for both FBDD and DEL,
high quality protein is essential for successful screens.
Covalent approaches
Covalent approaches to drug discovery
are becoming ever more acceptable as more covalent drugs are approved.
Understanding these in depth was the focus of Micah Niphakis (Lundbeck), who characterized
22 approved drugs containing 18 different warheads. The stability in buffer, liver microsomes,
and hepatocytes varied dramatically, though more recently approved drugs tended
to be more stable. Chemoproteomic studies revealed many off-targets in cells;
for example, all the kinase inhibitors tested hit BTK to some extent even when this was not
the primary target. The fact that the drugs are (mostly) safe and well-tolerated
is a useful reminder that just because we can detect something doesn’t mean it
is relevant.
Henry Blackwell described building
a 12,000-member covalent fragment library at AstraZeneca. Due to the presence
of a warhead, they relaxed
rule of three parameters, with MW ranging from
250-400 Da and ClogP from 0-4. Henry also discussed the successful use of this
library to identify covalent hits against the anticancer target BFL1 that were
optimized to k
inact/K
I ~ 7000 M
-1s
-1.
This accomplishment is all the more impressive given that screens using ASMS,
DSF,
19F NMR, and SPR had all failed to yield validated hits.
We recently
wrote about
electrophilic MiniFrags, and György Keserű (Research Center for Natural
Sciences, Hungary) described screening these against HDAC8 and the main protease
from SARS-CoV-2. He also mentioned that the set is available for purchase from
Enamine, so you can try it yourself against your favorite target.
As covalent modifiers become more
common we will see new metrics for characterizing them, as illustrated by
Benjamin Horning’s (Vividion) presentation, “Ligand Efficiency Metrics in
Covalent Drug Discovery.” He described Ligand Reactivity Efficiency (LRE),
defined as pTE
50(target, 1 hr) – pTE
50(glutathione, 1 hr),
where TE is the target (or glutathione) engagement. LRE is analogous to
LLE but focused on reactivity rather than lipophilicity. Despite my
post last week, the
metric could be useful, and I look forward to seeing what
Dr. Saysno and
friends will make of it.
Most covalent modifiers bind to a
target and remain intact, but Nir London (Weizmann Institute) has developed
Covalent Ligand Directed Release (CoLDR), in which a portion of the small
molecule leaves; applications include release of fluorescent or
chemiluminescent probes. Useable warheads include α-substituted methacrylamides
and sulfamate acetamides.
Although more recent covalent
drugs have targeted cysteine residues, there is growing interest in other amino
acid side chains. Nir mentioned that thio-methacrylate esters can react with lysine
residues, thought the kinetics are slow. And Carlo Ballatore (University of California
San Diego) described hydroxy-naphthaldehyde fragments that bound reversibly to a
lysine on the vascular target KRIT1.
Both plenary keynote speakers focused
heavily on covalent chemistry. Dan Nomura (UC Berkeley) described using chemoproteomics
approaches to find covalent molecules that could inhibit, degrade, or change
the cellular localization of myriad proteins.
Finally, K. Barry Sharpless (Scripps),
one of only five people to have been awarded the Nobel Prize twice, gave a rich description of sulfur (VI) fluoride exchange chemistry
(SuFEx), which included drawing chemical structures on a flip chart. He presented
the discovery of a fluorosulfate that is bactericidal against multiple resistant
forms of Mycobacterium tuberculosis. Interestingly, the molecule works
by modifying a catalytic serine residue which then cyclizes to form a β-lactam.
His passion for chemistry is obvious, but he also has personal reasons for pursuing
the second most deadly infectious disease: his brother died of tuberculosis before
effective drugs were developed. And with the rise of extensively drug resistant
TB, we’ll need new ones.
I’ll end on that note, but
please leave comments. And mark your calendar for April 14-17 next year, when
DDC returns to San Diego.