The three members of the inositol
hexakisphophate kinase family are potential targets for a wide range of
diseases, from Alzheimer’s to cancer to metabolic disease. However, current
inhibitors are not specific for individual isoforms. Also, the most potent
compounds contain a carboxylic acid moiety, which is usually at odds with brain
penetration. In a new ACS Med. Chem. Lett. paper, James Barrow and collaborators
at Johns Hopkins School of Medicine, the Lieber Institute for Brain
Development, and AstraZeneca describe neutral, selective inhibitors.
The researchers started with a
high-throughput biochemical screen of 17,000 fragments, each at 100 µM, against
IP6K1. The library itself is available here. After dose-response follow-up
studies, 90 hits confirmed, with IC50 values as good as 2 µM. Most
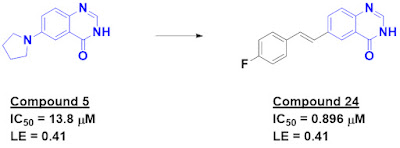
of the hits contained carboxylic acids, but compound 5 did not, and also had
good ligand efficiency.
No crystal structures of IP6K1
have been reported, so the researchers used an AlphaFold model for docking
compound 5. This suggested a fragment growing approach. A variety of
replacements for the pyrrolidine were attempted, and while some of these had
improved activity many also proved to be chemically unstable. Removing the nitrogen
and growing led to compound 24, which was both chemically stable and had sub-micromolar
activity.
The quinazolinone core itself was
associated with poor solubility, and the researchers made multiple attempts to
modify it, such as introducing additional nitrogen atoms or methylating to
remove a hydrogen-bond donor. Unfortunately, all these modifications led to significant
losses in potency.
Compound 24 is highly selective
for IP6K1 over IP6K2 and somewhat selective over IP6K3. Unfortunately, it
showed no cellular activity, possibly due to modest biochemical potency and
solubility. Nonetheless, this brief paper illustrates that starting with a
larger than normal fragment library can lead to new chemotypes. Screening the
larger library gave the researchers more chances to find fragments that did not
contain carboxylic acids. Indeed, the difficulty of modifying the quinazolinone
moiety demonstrates the utility of screening more molecules. Had the closely
related molecules been in the library, they might not have turned up as hits, but their
presence would suggest that relevant chemical space had been interrogated.
The paper is also a nice example
of optimizing hits in the absence of structural information. Although much
needs to be done to turn compound 24 into a chemical probe, the fact that it is
still so small (almost rule-of-three compliant) provides hope that this can be done.



No comments:
Post a Comment