Last week’s post focused on
fragment screening against RNA, and we continue the theme this week with a
paper published in Proc. Nat. Acad. USA by Kevin Weeks and collaborators
at University of North Carolina Chapel Hill, New York University, and
Université de Sherbrooke.
The researchers developed a
screening technology called SHAPE-MaP (Selective 2’-Hydroxyl Acylation analyzed
by Primer Extension and Mutational Profiling). Essentially, RNA in the presence
or absence of potential ligands is treated with an acylating agent that reacts
with the 2’-hydroxyl group on ribose subunits. This addition requires the
hydroxyl groups to be exposed, so ligands that bind in the vicinity may
directly block or cause conformational changes to change the patterns of
acylation. Conveniently, acylation causes mutations when the modified RNA is
sequenced, making modified sites easy to detect. Moreover, by clever uses of
“barcodes” in other regions of the RNA, multiple samples can be pooled and
analyzed.
Because the approach uses
sequencing to identify binding sites, long strands of RNA can be tested. In
this case, the researchers built an RNA construct containing a ‘pseudoknot’ structure
in the dengue virus genome as well as a thiamine pyrophosphate (TPP)
riboswitch, which changes conformation when it binds to TPP. (We wrote about a
different fragment screen against this riboswitch back in 2014). A set of 1500 rule-of-three
compliant fragments from Maybridge was screened, resulting in 41 hits. These
were then rescreened in triplicate, which winnowed the field to just eight
fragments, of which seven bound TPP and one appeared to be nonspecific. All
eight were assessed by isothermal titration calorimetry (ITC), which produced
measurable affinities for six.
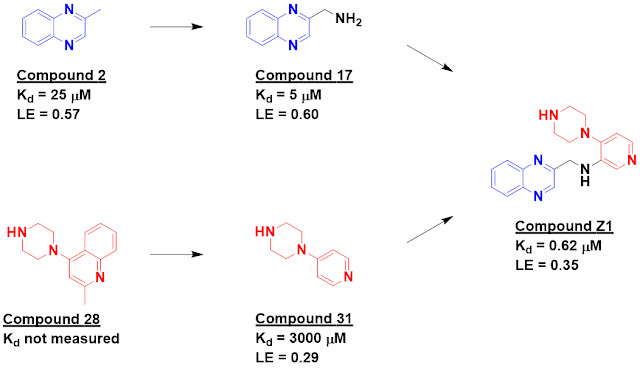
Compound 2 was the most potent
hit, and when the researchers tested 16 analogs they found some, such as
compound 17, with improved affinity. With an eye towards fragment linking, they
took a conceptually similar approach to SAR by NMR by rescreening the original
1500 fragments in the presence of compound 2 to look for ligands that would
bind at a second site. This yielded five hits, including compound 28. ITC
characterization of a more soluble analog, compound 31, revealed that it had
weak but measurably improved affinity in the presence of compound 2. A handful
of linked analogs were made, and while most of these had affinity similar or
worse than the best initial fragment, compound Z1 bound with sub-micromolar
affinity as assessed by ITC.
The natural ligand TPP binds to
the riboswitch with a Kd of 110 nM, and in doing so blocks in vitro
transcription of bound RNA. Despite having a similar affinity as TPP, compound
Z1 was much less effective at blocking transcription. Unfortunately, although
the researchers were able to obtain crystal structures of several molecules
bound to the riboswitch, including compound 17, they were unable to obtain one
with compound Z1.
This is a rare example of
fragment linking on RNA, and although the linked molecule does not show fully
additive affinity, it does have reasonable ligand efficiency. But like the example
last week, this paper illustrates how difficult discovering RNA binders is likely
to be. The confirmed hit rate is less than 0.5%, and this is for an RNA sequence that evolved specifically to bind low molecular weight
ligands. As the researchers note, none of the fragments bound the dengue virus
pseudoknot. Perhaps most RNA is truly undruggable, at least with small
molecules.



No comments:
Post a Comment