Once considered a time-intensive method justified
only by the most-promising molecules, crystallography is fast becoming a
routine technique for general fragment screening. Just last month we highlighted
an example where crystallographic hits were successfully used in virtual screens.
In a new J. Med. Chem. paper, Jeffrey St. Denis, Benjamin Cons, and
colleagues at Astex describe a crystallographic screen with a covalent fragment
library.
Driven in part by the rapid
development of KRASG12C inhibitors such as sotorasib, covalent
fragments are continuing to increase in popularity. To screen, you first need a
library, and here the researchers assembled a set of 114 molecules, most of
which were commercially available from Enamine. The majority (101) contained an
acrylamide moiety, the most common warhead in approved covalent drugs. The
library is rule of three compliant and fairly shapely, as assessed by deviation from planarity. Laudably, the structures of all 114 library members are
disclosed in the supporting information.
In addition to physicochemical
properties, the library was also characterized experimentally. Importantly, it
seemed to be quite stable when stored frozen at -80 °C in deuterated DMSO for
up to a year. Most (83%) of the compounds were soluble to at least 3 mM in aqueous
buffer. And just over half of the molecules withstood a test for reactivity
with 5 mM glutathione, the main nucleophile found in cells, assessed as >90%
remaining after 30 minutes at 37 °C.
As a test case, the researchers screened
the kinase ERK2, an oncology target for which Astex previously developed the
non-covalent clinical compound ASTX029. Soaking ERK2 crystals in 50 mM of each
fragment led to 29 bound ligands, a whopping 25% hit rate. Of these, 16 compounds
bound to C166, a cysteine near the hinge region in the active site. Unsurprisingly,
less reactive fragments made hydrogen bonding interactions with the protein,
while the more reactive fragments did not and also had poor electron density,
suggesting non-specific alkylation. As we noted in 2014, distinguishing
specific from non-specific binding is a general challenge when screening
irreversible fragments.
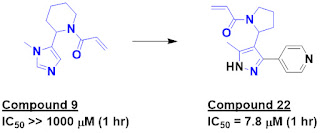
Among fragments making specific
interactions, compound 9 was chosen for further study due to its low inherent
reactivity and interesting structure. The molecule showed no inhibition of the
protein after 1 hour at 1 mM, but scaffold hopping and fragment growing led to
compound 22, with low micromolar inhibition after 1 hour. (A back of the envelope
calculation suggests kinact/Ki ~ 25 M-1s-1,
though I wish the researchers had reported this.)
This is an interesting paper, and
it is useful to get structural information up-front, but I can’t help thinking
that it would be more efficient to use an alternative method for the initial
screen. As the researchers acknowledge, many covalent fragment screens use
intact-protein mass spectrometry. For example, in a paper we highlighted in
2019, nearly 1000 fragments were screened against ten different proteins.
Moreover, as we noted last year, the ability of crystallography to find such
very weak hits can be a distraction as much as a blessing.
The current paper promises to “report
on the further developments and observations pertaining to our electrophilic
library in due course.” I look forward to seeing more.



No comments:
Post a Comment