Wnt proteins are implicated in a
variety of diseases, from Alzheimer’s to colorectal cancer. The enzyme Notum shuts
down signaling by removing a palmitoyl group from Wnt. Last year Practical
Fragments highlighted several series of Notum inhibitors identified from biochemical
and crystallographic fragment screens. The researchers behind those efforts, including
Paul Fish and Fredrik Svensson (University College London), have now published
a successful virtual screen against the enzyme in J. Med. Chem.
Starting with 1.5 million
compounds available from ChemDiv, the researchers chose 534,804 based on a variety
of computational filters including molecular weight (200-500 Da), number of
hydrogen bond donors (<=2) and ClogD (-4 to 5). A virtual screen of these
(using Glide) produced 1330 high-scoring hits, of which 1088 were chosen for
purchase. Of these, 952 were available, a much higher percentage than the
ZINC15-reliant paper we wrote about earlier this year.
All 952 compounds were tested in
a biochemical assay, and the 44 that gave >50% inhibition at 1 µM were then
tested in dose-response format. This yielded 31 compounds with IC50
values < 500 nM. These could be subdivided into four structurally related
clusters and eight singletons. Further triaging removed compounds likely to
cause assay interference as well as those similar to known Notum inhibitors.
This left two clusters and two singletons.
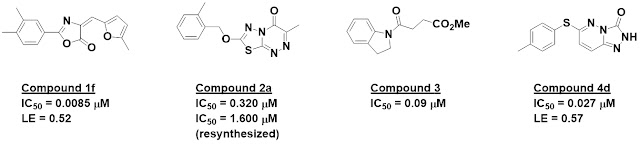
Compound 1f was the most potent
member of a series of 9 related (and possibly covalent) inhibitors. Although these
strongly inhibited the enzyme in the biochemical assay, they were essentially
inactive in a cell-based assay. They were also highly insoluble and showed low
cell permeability, and were thus dropped.
Compound 2a was one of two
related molecules that were also quite potent when initially tested.
Unfortunately, when the molecules were resynthesized they turned out to be significantly
weaker and were also not very soluble, so this series was also halted.
The singleton compound 3 turned
out to be a covalent inhibitor; the catalytic serine formed an ester with the
molecule. The mechanism is more fully described in this open-access J. Med.
Chem. paper.
That leaves the second singleton.
Compound 4d was not just active in the biochemical assay, it also showed
sub-micromolar cell activity. SAR, guided by crystallography, ultimately led to
low nanomolar inhibitors. The pKa of compound 4d was measured to be
7.9, which is less acidic than many previously reported Notum inhibitors and thus
more likely to be cell permeable. This turned out to be the case experimentally,
and the compound was also stable in mouse liver microsomes. Pharmacokinetics in
mice were promising for several compounds, but unfortunately brain penetration
– which the researchers were hoping for – was negligible. (This could be an
advantage for peripheral diseases.)
This is a nice
example of lead discovery in academia. Like last week’s post, it also illustrates
that fragments themselves can be quite potent. Indeed, although the researchers
were looking for molecules up to 500 Da in their virtual screen, all of the
best hits were fragment-sized. Another illustration that small is beautiful.



No comments:
Post a Comment