DOS – diversity-oriented synthesis – intentionally generates a disparate set of compounds from a small number of starting materials in just a few synthetic steps. The idea is that if
any of these turn out to be hits, it will be straightforward to make analogs. Since
figuring out what to do with a fragment is a common bottleneck, DOS-derived
fragments could help. An open-access paper published in Chem. Sci. by David
Spring (University of Cambridge) and collaborators from several institutions
demonstrates how to use DOS to move fragment hits forward.
The researchers had previously
disclosed (also open access) a rule-of-three-compliant DOS library of 40
compounds derived from racemic α-methyl propargylglycine. In the current paper,
these molecules were screened crystallographically at 500 mM on the Diamond
Light Source XChem platform against three protein targets.
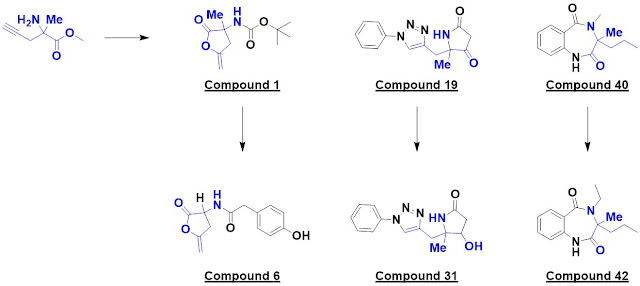
The first, penicillin binding
protein 3 (PBP3) from P. aeruginosa, is a classic antibiotic target. A
single hit, compound 1, was identified. Interestingly, this turned out to be a
covalent modifier, with the catalytic serine opening up the lactone. The researchers
made 10 analogs exploring four different vectors, each in five synthetic steps
using cheap reagents (< £3 per gram). These were screened crystallographically
and six bound; one example is compound 6.
The next protein screened,
cleavage factor 25kDa (CFI25), is a subunit of the pre-mRNA cleavage
factor Im. (No, I hadn’t heard of it either.) A crystallographic screen yielded
two hits, one of which – compound 19 – was elaborated into 14 analogs. This provided
some preliminary SAR around the phenyl ring as well as a surprise: compound 31
bound to a different region of the protein.
Finally, the researchers screened
activin A, a member of the transforming growth factor β superfamily. Compound
40 was elaborated into 14 analogs exploring four vectors, and compound 42 was found
to bind in a similar manner.
There are several take-aways from
this paper. First, DOS libraries can be remarkably diverse: compounds 1, 19,
and 40 are all quite different from one another. Second, although I hesitate to
discuss hit rates from such a small library, it is encouraging that hits were found
at all even from fairly shapely fragments. These are also the first reported
small molecule binders for CFI25 and activin A. Laudably, all the
structures have been deposited in the protein data bank, extensive details are provided in 185 pages of supplementary material, and the library itself
is available for screening at XChem.
One downside to crystallographic
screening is that affinities are not part of the package, and some hits may be so
weak as to be difficult to advance. But the researchers note they are further
characterizing the compounds in the hope of producing more potent analogs. Although
Teddy’s deadline for demonstrating a highly ligand-efficient molecule from DOS has
long passed, hopefully the Safran Zunft Challenge will soon be met.



2 comments:
What does "vector" mean in this context?
Good question. Vectors are the regions of the molecule suitable for diversification. For example, in the case of compound 40, the four vectors include the N-methyl, the methyl and ethyl substituents off the stereogenic carbon, and the amide at the "bottom" of the molecule. Each of these was varied independently to make 14 analogs.
Post a Comment