t
t r
r -sp
-sp k
k t
t v) adj.
v) adj.
1. Looking back on, contemplating, or directed to the past.
2. Looking or directed backward.
3. Applying to or influencing the past; retroactive.
I would add: 4. Looking back on the past, to influence the future.
In this vein, a recent paper by Ferenczy and Keserű in J. Med Chem looks back on hit-lead optimizations derived from fragment starting points. In this very interesting paper, they look at 145 fragment programs and evaluate the properties of the original hit and then again as it progresses into the lead. Of the 145 programs, these were aimed at 83 proteins of which 76 are enzymes, 6 are receptors, and 1 is an ion channel. These programs evolved into leads, tools, and clinical candidates. The authors set out to answer three questions: 1. do fragments eliminate the risk of property inflation, 2. how do ligand efficiency metrics support fragment optimizations, and 3. what is the impact of detection method, optimization strategy, and company size on the optimization.
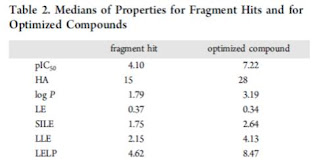
Table 2 shows the median the calculated properties for the hits and optimized compounds. The pIC50 improved by roughly three orders of magnitude, but ligand efficiency (LE) stayed roughly the same. Log P increased but SILE did not. SILE was a metric I was not familiar with and is calculated by pIC50/(HAC)^0.3. SILE is a size-independent metric of ligand efficiency. I won't attempt to reproduce all the graphs they generated; get the paper. Some interesting data points: the median fragment hit had 15 heavy atoms (see related poll here), the median size of leads is 28 heavy atoms, and good fraction of hit-lead pairs changed less than 5 heavy atoms (which of course is well known from here). So, what is the answer for their first question? If you are looking at something like LE, then these hit-lead pairs maintain the efficiency. If you are looking at logP, then the answer is no, the hit-lead pairs get greasier. I would really like to see more granularity here (see What's Missing below). Their major comment here is that SILE and LELP (work by the author's previously reviewed by Dan) are the two best metrics to monitor as hit-lead optimization is underway. Increases in both metrics correlate with increases in FBDD programs.
They then looked at the screening method. The breakout of primary screening (in their definition the first one listed when multiple methods were used) was 38% biochemical, 25% NMR, 18% X-ray, and 11% virtual. This is an interesting contrast to these results; SPR is not the dominant screening technique (8% tied with MS). So, does this mean, >40% of practitioners are using SPR, but as a secondary screen?
Table 4 then does a pairwise comparison of the metrics based upon primary screen origin of the fragment (see What's Missing below). Biochemical screens yield the most potent hits (4.75) while NMR (3.53) has the least potent. X-ray has the smallest hits( 13 heavy atoms) while virtual screening the largest (17 HA). I don't think any of this is surprising; the authors point out that hit properties exhibit a significant dependence on the method used. It is noted that optimization tends to diminish differences in hit properties. Again, I think this is not surprising; thermodynamics and medchem are all the same no matter how big or small the molecule. They do point out that biochemical based hits preserve their advantage after optimization, primarily relative to NMR. They posit that the difference is that more potent compounds need less "stuff" to become sufficiently potent, and thus have a better mix of medchem for potency and other property optimization. Weaker starting points need more bulk, more atoms, to become equipotent with biochemical starting points, they suggest. Lastly, they show that structural-based optimization efforts are better than those without structural information. The structural information comes from X-ray primarily (52%) and NMR (10%). Interestingly, they reference this poll from the blog on whether you need structural information to prosecute fragments. [As an aside, since I wrote that blog post and it was reference in a paper, do I get to add it to my resume?]
Finally, they break the originating labs into three categories: academic (18%), small/medium enterprises (SME)( 37%), and Big Pharma (45%). The SME results are superior to those achieved by the academics and Big Pharma. Their explanation is that SME's tend to be more platform focused and predominantly ensure "structural" enablement of targets. 75% (40/53) optimizations at SMEs used protein structural information, while "only" 62% of those at Big Pharma did. They do not rule out the differences in target selection at those two different groups of companies. I would like to propose an alternative hypothesis (tongue-not-entirely-in-cheek): Big Pharma has "old crusty" chemists who don't understand fragments and thus just glom on hydrophobic stuff to increase potency because "that's how we always do it" while SME have innovative chemists. And of course academia is just making tool compounds and crap.
One thing that I would like to emphasize is that Ro3, metrics, and so on should not be used as hard cutoffs. As shown in Figure 11, even compounds that are outside the "preferred" space can reach the clinic. The best way to view them is akin to the Pirate Code; they are more guidelines than rules.
What is Missing? The supplemental information (which the authors are willing to kindly share) does not break out the targets into specific classes. However, they do list each target, so it should be easy to add this a data column. More importantly, they do not break out hit-lead pairs into those that were optimized for use as tools, clinical candidates, and leads (and which of the leads died). Tools are never supposed to look like leads (but you are lucky if they do), so their inclusion here can be biasing the results. Although, it is likely that the of the 145 not very many of the examples are strictly tools; it would be nice to know though.
I am struck by the information denseness of Table 4 and wish that instead of pairwise comparisons, they had instead simply list the hit-lead metrics for each methodology. I think there is gold to be mined in Table 4 and just like real gold is hard to find.
I have not addressed every single point made by the authors. I, for one, am hoping that they will continue their analyses (especially with an eye to some of What is Missing). I hope that there will be a significant amount of discussion around these points. I will make sure we hit on this at the breakfast roundtable at the upcoming CHI FBDD event in SD (I even have the same pithy title as last year!)