Two years ago we highlighted what
was likely the largest crystallographic fragment screen against any target, the
macrodomain (Mac1) of the nonstructural protein 3 (Nsp3) of SARS-CoV-2. Mac1
dampens the cellular immune response to viral infection by removing ADP-ribose
from various proteins. Separate mutational studies suggested this enzyme could be a good
target for treating COVID-19.
The 234 fragment hits identified in 2021 could serve as good
starting points. This has proven to be true, as demonstrated in a paper just
published (open access) in Proc. Nat. Acad. Sci. USA by Brian Shoichet,
James Fraser, and collaborators at University of California San Francisco,
University of Oxford, Diamond Light Source, Enamine, and Chemspace. Despite the wealth of fragment
hits, none of them were particularly potent; the best had an IC50
value of 180 µM in a homogenous time-resolved fluorescence (HTRF) competition
assay. In the new paper, the researchers leveraged computational methods to advance these fragments.
First, they explored a fragment-linking
approach termed Fragmentstein. This entailed choosing pairs of fragments
that bound in close proximity to one another, merging or linking them, docking
them to ensure the new molecule would bind in a similar manner to the component
fragments, and then searching make-on-demand libraries in Enamine’s REAL database.
Four pairs of fragments were evaluated, and 13 of 16 designed compounds were
synthesized. Eight of these confirmed crystallographically, and two showed low
micromolar activity in the HTRF assay. Interestingly, both of these came from the
same fragment pair, ZINC922 and ZINC337835. The best molecule was a mixture of
diastereomers, and one of the pure stereoisomers turned out to be submicromolar.
The potency of this fragment is
all the more impressive given the low affinity of the initial fragments, which could
only be crystallographically characterized using PanDDA, a method to find
low-occupancy ligands that we wrote about here. Unfortunately, the
compound has low cell permeability, likely due to the carboxylic acid moiety.
In addition to the linking
approach via Fragmentstein, the researchers also conducted two virtual docking
campaigns with more than 400 million molecules with molecular weights between
250-350 amu. Of 124 molecules purchased and tested, 47 confirmed
crystallographically and 13 confirmed by HTRF, with IC50 values from
42 to 504 µM. (Ten of the 13 HTRF hits were also crystallographic hits. The
researchers suggest the difference in confirmation rate is due at least in part
to compound concentrations, which were 10-40 higher in the crystallographic
screens.) In general the crystal structures confirmed the computationally predicted
binding modes, particularly for fragments with measurable activity. Structure-based
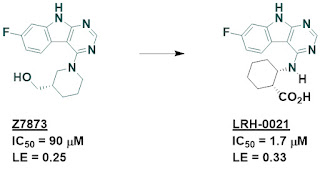
optimization of some of these fragments led to multiple low micromolar inhibitors,
such as LRH-0021. Despite the carboxylic acid, this molecule is cell permeable.
This paper nicely illustrates how
even very weak fragments can lead to multiple and very different series of
inhibitors. The researchers acknowledge that the molecules are still at an
early stage of development; indeed, they note that there are currently no good
cellular assays to even assess the effect of Mac1 inhibition. Laudably, all the
structures are deposited in the Protein Data Bank, which should provide a useful
resource not just for further efforts on this protein but for understanding
molecular interactions more generally.




No comments:
Post a Comment