Three years ago we discussed using DNA-encoded libraries to find and link fragments. In a new
open-access Bioorg. Med. Chem. article, Nicolas Winssinger and colleagues
at University of Geneva report a different version of this approach.
Rather than using DNA, the researchers
constructed their libraries with peptide nucleic acids (PNAs), which can be assembled
using traditional solid-phase peptide synthesis and will also hybridize to DNA.
Each PNA is coupled to a different fragment, and the fragment-PNA molecules are
then bound to microarrays of DNA such that two fragment-PNA molecules bind to a
single DNA strand. In this case the researchers used 250,000 combinations of
fragment pairs.
Next, a protein of interest (here
the anti-apoptotic cancer target BCL-xL) was screened at 50 nM. Binding
to specific pairs of fragments was assessed by fluorescent detection of the
protein at various spots on the microarray.
Trying to figure out which of the
fragments are best – and how to link them – is “not trivial,” so the researchers
took a combinatorial approach. Based on the first screen, they generated a new library of 10,000 molecules in which 10 sulfonamide-containing
fragments were linked to 100 heterocycle-containing fragments using 10
different linkers. These compounds were screened using the same microarray technology,
and the best binders were then resynthesized with a biotin tag rather than the
PNA.
The biotinylated molecules were
able to pull down recombinant BCL-xL in solution. Two of them,
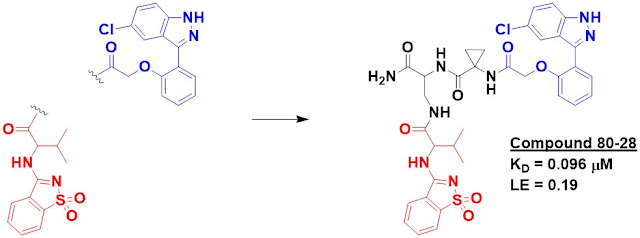
including compound 80-28, were even able to pull down recombinant protein that
was spiked into cell lysate. Importantly, neither fragments 80 nor 28 did this by themselves. The affinity of 80-28 was measured by SPR to be 96 nM.
Finally, the researchers tested 80-28
in K562 cells and found that it was cytotoxic with EC50 = 1.7 µM.
They compare this favorably to venetoclax, the second fragment-based drug to be
approved. However, this is a disingenuous comparison: venetoclax was specifically designed to
bind less tightly to BCL-xL than to the related protein BCL-2. A more
appropriate comparison would be a molecule such as navitoclax or the specific BCL-xLbinder A-1155463.
Like most BCL-family binders, compound
80-28 is also a rather unusual looking molecule, with a high molecular weight
of 735. Unlike navitoclax and venetoclax, it also has 7 hydrogen bond donors
and many more rotatable bonds. The long floppy linker in particular is
something the earlier DNA-based fragment-linking work sought to fix. As we
noted then, such linkers may be an inherent liability with the approach. From a technology
perspective this is interesting work. But from a drug discovery perspective it
still has some way to go to prove itself practical.



No comments:
Post a Comment