Advancing fragments without high-resolution structural information
remains a challenge scientists often choose not to take on, according to our
poll last year. But for many appealing targets, such as membrane proteins,
structural information is difficult to obtain. In a new paper in J. Med. Chem., Peter Kolb and
collaborators at Philipps-University Marburg and Vrije Universiteit Brussel
describe a computational strategy.
The approach, called “growing via merging”, starts with a
core fragment that binds to a target, in this case the β2-adrenergic receptor (β2AR). Ideally this interaction is structurally
characterized, but if not a model can suffice. Here, the researchers started with
five fragments they had previously discovered. All of these had in common a
lipophilic core with a primary or secondary amine appendage; this is a known
pharmacophore for β2AR, so
modeling could be used to orient the fragments.
Next, this core fragment is derivatized in silico with other
fragments using a selection of 58 common reactions. Since all five fragments
contained an amine, reductive amination was used here. A set of nearly 19,000
fragment-sized aldehydes and ketones was extracted from the ZINC database and computationally
transformed into amines – as if they were reacted with one of the core
fragments. These were then docked into the receptor, and those that did not
overlap with the core fragments and also placed the amine near the amine of the
core fragment were kept for further analysis.
The top 500-scoring fragments were then “reacted” – again in
silico – with the core fragments and again docked. Eight of these were actually
synthesized and tested for binding, of which four had higher affinity than the initial
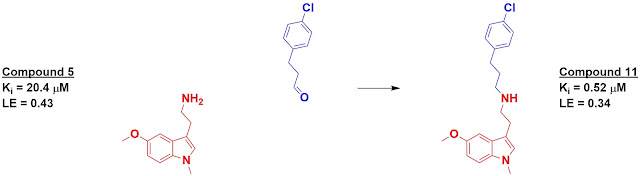
fragments. The best, compound 11, showed a 40-fold boost in affinity over its
starting fragment.



3 comments:
Just wondering:
"These were then docked into the receptor, and those that did not overlap with the core fragments and also placed the amine near the amine of the core fragment were kept for further analysis."
Shouldn't it read:
"These were then docked into the receptor, and those that did overlap with the core fragments and also placed the amine near the amine of the core fragment were kept for further analysis."
Hi Markus,
Good question - it's a little tricky as they actually want to make sure the fragments don't bind in the same location as the core fragment with the exception of the amine. In the example shown, the blue aldehyde would be converted to a primary amine; the hope is that the chlorophenyl moiety would not bind in the same location as the indole of compound 5. You are correct that in the following step the indole moiety of compound 11 should bind in the same location as the indole of compound 5.
Hi Markus and Dan,
Dan correctly described the procedure.
What I want to clarify is that we are basically doing a fragment merging procedure between the core-fragment and a given surrogate (i.e. the building block we attach). Thus we don't want them to clash against one another (we assume the merging will fail).
As you know fragments are very small, so they can bind everywhere. In computational approaches such as docking, this is translated by a high number of possible poses (i.e. predicted binding modes). In our case, a single surrogate can have up to thousands of poses. Even if we limited the binding site for docking (in this case in the SBP), some of the surrogate poses will clash with the core fragment, this is why they need to be post-filtered (figure 5 described it well: https://goo.gl/FZah2c).
A more general workflow is also available in the SI (https://goo.gl/RziBgH), fig S1.
I hope I could bring some light here. Please let me know if you need more details, I will happily provide them.
Florent
P.S: I am the main author
Post a Comment