One of the more challenging
protein-protein interactions targeted by drug hunters is the interface between
the transcription factor NRF2 and its repressor KEAP1. This is part of the
cellular defense against reactive oxygen species; increasing NRF2 activity may
be useful for treating a variety of diseases. Unfortunately, the binding site
on KEAP1 that interacts with NRF2 is large and has a predilection for
carboxylic acids. Thus, many of the molecules reported as inhibitors tend not
to be druglike. Anders Bach (University of Copenhagen) and a multinational team
of collaborators sought to do better, and have just published some of their journey
in J. Med. Chem.
The researchers had previously tested
19 reported small-molecule KEAP1 inhibitors, of which only nine confirmed. (This
is a salutary reminder to take any individual publication with a large grain of
salt.) The nine fell into six chemical series (two shown below), and the researchers decided to fragment
some of these molecules into 77 fragments. The fragments were then tested in
four assays: fluorescence polarization (FP), a thermal shift assay (TSA), saturation
transfer difference (STD) NMR, and surface plasmon resonance (SPR).
Primary hit rates were generally
high, from 25%-64%, but long-time readers will not be surprised that the
overlap was not great: no fragments hit in all four assays, and only eight hit
in three. As the researchers point out, this could reflect differences in
sensitivity, conditions (from 3-8% DMSO and from 0.5 to 8 mM fragment), and different
types of false positives and false negatives. Interestingly, and in contrast to
previous work, overlap was good between STD NMR and SPR.
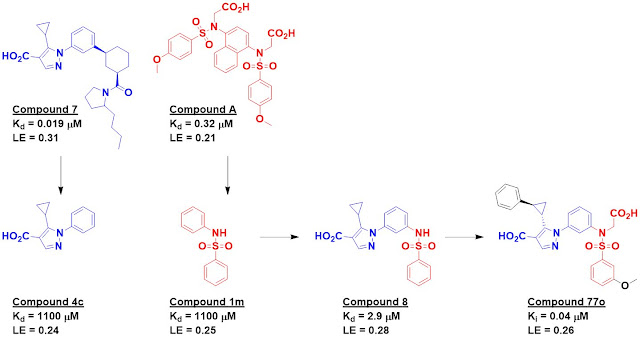
Crystal structures of seven hits
were solved bound to the protein, and compounds 4c and 1m (from different
precursor molecules) were merged to provide compound 8, with low micromolar
affinity. Compound 8 was the subject of considerable medicinal chemistry, with
five different vectors chosen for growing. Despite being structurally enabled,
the researchers struggled; changes that improved affinity in one context did
not do so in another. After considerable effort, the researchers obtained
compound 77o, with mid-nanomolar activity.
Compound 77o is stable in human
plasma and mouse liver microsomes. Unfortunately, and unsurprisingly given the two
carboxylic acids, it has poor permeability. Indeed, a fragment-derived KEAP1
inhibitor we described previously has only a single carboxylic acid, as does
precursor compound 7. As the researchers themselves acknowledge, “the physicochemical
properties of our compounds are not favorable for membrane permeability.”
Nonetheless, this paper is a lovely
example of fragment-based deconstruction reconstruction (FBDR) and is well
worth studying for the thorough descriptions of fragment screening in orthogonal
assays and structure-based design. Another lesson may be that despite considerable
effort, the final molecule is far from a chemical probe, let alone a drug. Perhaps
some targets truly are undruggable. Or maybe – as for other seemingly undruggable
targets – a change in strategy is needed.
I have seen the benzo-sulfonamide coming out fragment screening many many times, later on suffering from HSA binding degradation or permeability issues.
ReplyDeleteHi Dan. Thanks for posting our paper. After reading this blog for many years it is great to see our work mentioned.
ReplyDeleteHi Unknown. Thanks for the good point. We also see that compound 8 (with one acid only but with the sulfonamide) has low permeability (Fig S4 and Table S4). So the permeability issues could just as well be due to that...a combo I guess, at least it is not all about the acid(s). 8z is also low in permeability (amide instead of acid and with a sulfonamide).
BR
Anders
Thanks Dan, I love the concept of peeling back a dubious starting hit (cmpd A) to get to a more palatable fragment. What's the best review in your opinion on fragment-based deconstruction reconstruction (FBDR)? I'm curious to see what other kinds of fragments have been "recovered" in this way from bigger hits. Is there a solid review covering different strategic approaches to FBDD?
ReplyDeleteCheers,
Dennis
Hi Dennis,
ReplyDeleteYes, deconstruction can be a powerful approach. The only dedicated review of which I'm aware was published in 2014:
https://pubmed.ncbi.nlm.nih.gov/25263697/
I've highlighted more recent examples on Practical Fragments, most of which can be found in this search:
http://practicalfragments.blogspot.com/search?q=deconstruction&max-results=20&by-date=true
And these approaches are also called out in our annual fragment-to-lead perspectives, the most recent of which can be found here:
https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c01608
Enjoy!
Dan