Practical Fragments
recently introduced WTF as a light-hearted jab at the continuing proliferation
of metrics to evaluate molecules, but there is an underlying problem: which
ones are useful? In a provocative paper just published online in Bioorg. Med. Chem. Lett. (and also discussed over at In the Pipeline) Michael
Shultz asks:
If one molecular
change can theoretically alter 18 parameters, two shapes, the rules of 5, 3/75,
4/400 and ‘two thumbs’ while simultaneously affecting at least nine composite
parameters and countless different methods of representing data, how is a
practicing medicinal chemist to know if any specific modification was actually
beneficial?
Shultz focuses on three parameters in depth: ligand efficiency (LE), ligand-efficiency-dependent lipophilicity (LELP), and lipophilic ligand efficiency (LLE, also referred to as lipophilic efficiency or LipE). He
conducts a number of thought experiments to see how these metrics change when,
for example, a methyl group is changed to a t-butyl group or a methyl sulfone.
He also examines how the metrics perform against historical data from Novartis
lead-optimization programs.
One problem with LE is that, although it was introduced to
normalize potency and size, it is still highly dependent on number of heavy
atoms (heavy atom count, or HAC): addition of one atom to a small fragment will
have a more dramatic effect on LE than addition of one atom to a larger
molecule. This has led to metrics in which larger molecules are treated more
leniently, but because of the way all these metrics are mathematically defined,
none achieve completely size-dependent normalization.
More seriously, LE ignores lipophilicity, which seems to be
correlated with all sorts of deleterious properties. With a nod to Mike Hann’s
“molecular obesity,” Shultz notes that the widely used body mass index (BMI)
“cannot distinguish between the truly obese and professional athletes of
identical height and weight. Similarly, HAC based composite parameters such as
LE cannot distinguish between ‘lean molecular mass’ and groups of real
molecular obesity.”
LELP addresses this shortcoming by incorporating clogP, but
it has problems of its own. For example, “the effects of lipophilicy are
magnified as molecular size increases.” More alarmingly, as clogP approaches
zero, LELP becomes increasingly insensitive to both size and potency; a
femtomolar binder would have the same LELP as a millimolar binder when clogP =
0.
In contrast to both LE and LELP, LipE (or LLE) is
size-independent, so a change in potency or lipophilicity will produce the same
change in LipE no matter the size of the initial molecule. Shultz uses data
from two lead optimization programs to show that LipE behaves better than LE or
LELP. This is in contrast to a previous report that suggested LELP to be
superior to LipE, albeit against a different data set.
Shultz further notes that LipE can be thought of as the
tendency of a molecule to bind to a specific protein rather than to bulk
octanol:
LipE = pKi - clopP = log [EI]/([E][I]) – log ([Ioctanol]/[Iwater])
where E stands for protein and I stands for inhibitor
Although this is a simple consequence of the math, it is a
nice way of visualizing an otherwise abstract number. Moreover, it suggests
that optimizing for LipE could optimize for enthalpic interactions, a topic
Shultz explores in depth in a companion paper.
Overall Shultz raises some excellent points, but I still
believe there is value in LE (and LLEAT), particularly in the
context of fragments, which usually have low affinity. Ligand efficiency can
prioritize molecules that might otherwise be overlooked. For example, it is
hard to get too excited over a 1 mM binder, but if the hit has only 8 heavy atoms it
could be valuable.
Turning
to my own miniature thought experiment, fragments 1 and 2 have very similar
LipE values, but the LE of Fragment 1 is better, and arguably makes a more attractive fragment hit.
Of course, in the end, rules should not be followed slavishly; the most lucrative drug of all time, Pfizer’s atorvastatin, violates Lipinski’s rule of five. Papers like this are important to highlight the problems and inconsistencies that underlie some of our metrics. Ultimately I’ll take biological data and the intuition of a good medicinal chemist over any and every rule of thumb.
What do you think? What role should LE, LELP, and LipE play in
drug discovery?
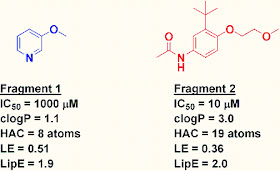
Dan, based on your hypothetical data I calculate an LE of 0.36 for Fragment 2, which would make it an OK starting point.
ReplyDeleteFor me, and I am sure I have said this before, LE is not meant to represent reality, which is what I think most metrics are trying to do. LE, OTOH, is a useful guide to help you decide if you are making smart, efficient use of chemistry space, rather than just glomming stuff on. I would never argue 0.29 is much worse than 0.31. I would say that 0.21 is much worse than 0.31 however.
ReplyDeleteUsing LE (pIC50/HAC) I get 3/8 =0.375
ReplyDeleteand 5/19 =0.26.
Thanks Anonymous, good catch - I've corrected it above. However, the LE of Fragment 1 is still better, which would flag it as interesting despite the pathetic 1 millimolar IC50. LE can be useful for flagging such fragments that might otherwise be overlooked.
ReplyDeleteDr. Teddy, try with LE = -RT*ln(IC50)/HAC ...
ReplyDeleteI defined my LE, why should I use yours?
ReplyDeleteThe problem is not with the papers or the studies (with the exception of the "Golden Ratio" paper). The problem is with implementation.
ReplyDeleteAll of these studies should aid the practicing medicinal chemists in how to think about things from multiple perspectives and focus on why structural changes result in activity differences, rather than "a methyl group made that number go up and the other number go down".
Unfortunately, some (many) are written in a way that suggests that chemists should stop thinking and just follow the rules.
Worth reading and thinking about, but not necessarily worth reading and acting upon.
Thanks for sharing such a great information.Am looking forward for your net post.
ReplyDeleteAnonymous got it right. In my view the Shultz paper is hot air as a result of not understanding why we use these metrics.
ReplyDeleteWe use these measures because us humans do not cope well with multi-objective optimisation. So we reduce space to two parameters 9potency & size, potency & lipophilicity).
You use the measure that suits your problem - that is why I don't buy Shultz's argument that "LLE is best" for mathematical reasons. It won't work when I am trying to decrease the size of my mols, and likewise LE will not work when optimising lipophilicity (d'uh!).