In templated or in situ reactions,
bonds form between two fragments that are brought together in the context of a
larger molecule such as a protein. We have written previously about dynamic combinatorial
chemistry (DCC), which depends on reversible bond formation where the larger
molecule shifts the equilibrium toward the linked fragments. For irreversible
bond formation, the larger molecule effectively catalyzes the formation of an
inhibitor (or at least a binder). In a recent Angew. Chem. Int. Ed. paper
Jyotirmayee Dash and colleagues at the Indian Association for the Cultivation
of Science describe an application of the latter that uses RNA as the template.
The target of interest was TAR
RNA, a short region of viral RNA essential for HIV replication. We have previously
highlighted a few examples of fragment screening against RNA (here, here, and
here), including TAR, but most of the hits were weak.
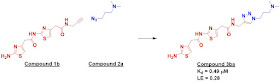
The researchers used azide-alkyne
cycloaddition, the quintessential click chemistry reaction. They built a small
library of four alkynes (only one of which was fragment-sized) and 11 azides.
All of these were incubated together (4 µM of each alkyne and 1 µM of each
azide) in the presence of 5 µM biotin-labeled TAR RNA for 72 hours. (The reaction
is typically slow at room temperatures unless catalyzed by metal ions.) Magnetic streptavidin-coated
beads were then used to capture the RNA and any bound ligands, which were identified
by HPLC-MS. Control experiments were run with TAR DNA or a mutant form of TAR RNA
lacking an essential bulge. The result was one fairly potent compound (below) that was
specific for TAR RNA, as well as a couple other molecules that were both weaker
and less specific.
The affinity of compound 3ba for
TAR RNA was measured by isothermal titration calorimetry (ITC) as well as by a
fluorescence assay, which were in good agreement. Importantly, the ITC data
suggested 1:1 binding, which is particularly important given that the ligand
contains two 2-aminothiazoles, a moiety that has been called a PrAT for its
promiscuous behavior. Finally, the ligand could displace the Tat peptide at low
micromolar concentrations, suggesting that it is binding at the biologically
relevant site of TAR.
I do have a few quibbles. It would
have been interesting if the researchers had reported the affinity of the azide
and alkyne themselves to see how much of a boost they got by linking. And since the most
potent molecule is not always selected from target-guided synthesis, it would
have been interesting to make and test other possible cycloaddition products to see if they missed anything useful.
Still, it is nice to see a submicromolar
RNA binder come out of an in situ screen. Targeting RNA with small molecules
has recently become trendy, and it will be fun to see how far approaches like
these can go.
Very interesting to read. Targeting RNA might be the evolution of drug discovery.
ReplyDeleteThe compound contains an amino-thiazole, well know for its extreme promiscuity. Though the binding to the target looks genuine, the promiscuity issue may cause trouble down the line.
ReplyDelete