As we recently noted, there are hundreds of human kinases in
the human genome, and figuring out what they do is not easy. Selective small
molecule inhibitors can probe an uncharacterized kinase’s function, but these
don’t exist for the majority of proteins. Such was the case for maternal
embryonic leucine zipper kinase (MELK), a potential anti-cancer target. In a
recent paper in ACS Med. Chem. Lett.,
Chris Johnson and collaborators at Astex and Janssen describe how they created
one.
The researchers started with a screen of the ~1500 Astex fragment library using both ligand-observed NMR and protein thermal shift
assays. Hits were soaked into crystals of MELK, resulting in “a large number”
of structures of fragments bound to the hinge-binding region. It is certainly
possible to develop selective inhibitors that bind to the hinge region, but doing
so is seldom straightforward, and thus the researchers were particularly
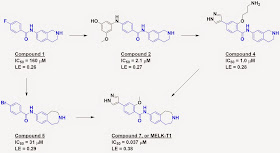
interested in unusual fragments. Compound 1 caught their attention because it
makes just a single hydrogen bond with the hinge region via the carbonyl oxygen
in the fragment; most other hinge binders form two or three hydrogen bonds.
Compound 1 was well-positioned for fragment growing, and the
addition of a phenol moiety (compound 2) led to a nice boost in potency and
maintenance of ligand efficiency. Crystallography revealed that the phenolic
oxygen was both a hydrogen bond donor as well as an acceptor, and replacing this
moiety with a pyrazole led to compound 4, with slightly better potency.
Pyrazoles themselves are often hinge binders (two of Astex’s clinical compounds, AT9283 and AT7519, contain them), but crystallography revealed that
the binding orientation remained the same as in the original hit.
Expanding the aliphatic ring of compound 1 by one methylene
gave compound 5, with a higher affinity and a slightly better fit to the
protein, and adding bits from compound 4 gave mid-nanomolar compound 7, or
MELK-T1. Impressively, the researchers improved the ligand efficiency of their
molecules even as they became larger.
Finally, the moment of truth: would optimizing a fragment
with an unusual hinge-binder lead to a selective inhibitor? The researchers
tested MELK-T1 in a panel of 235 kinases, and happily only 6 were inhibited
>50% at 1 µM [compound]. Notably, these did not include AMPK, which has 60%
identity to MELK in the kinase domain. MELK-T1 was also cell-permeable.
This is a classic example of FBLD enabled by a robust
protein construct suitable for crystal soaking. Getting to MELK-T1 required the
synthesis of only ~35 compounds, and should lead to some interesting biology.
At the same time, the researchers took a different approach to come up with
another series of leads, which will be the subject of my next post.
It is a nice piece of work and thanks for publishing. Given what the folks at Astex have been saying recently about their fragment library trending towards lower heavy atom counts and reduced complexity compound 1 stands out as being right up there with a 20 heavy atom count and two distinct pharmacophores.
ReplyDelete